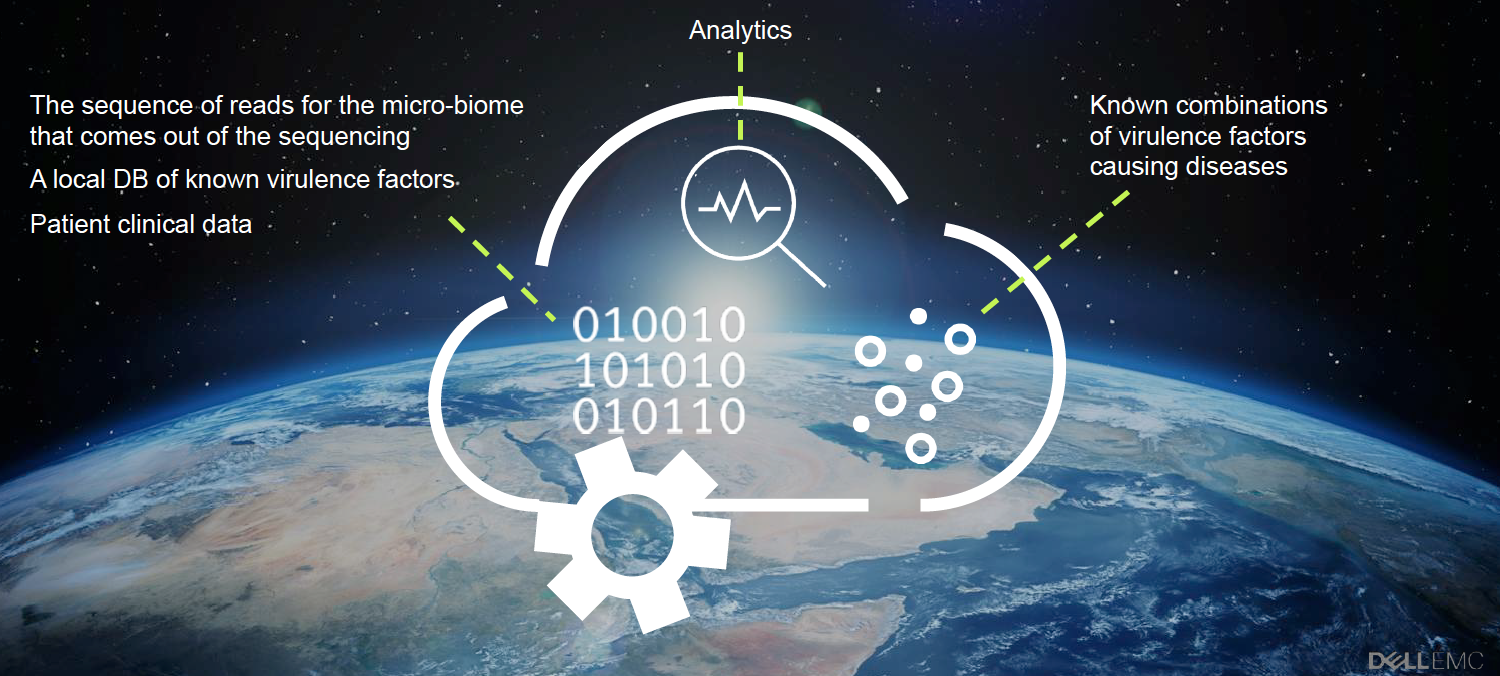
Analysis of very large genomic datasets has the potential to radically alter the way we keep people healthy. Whether it is quickly identifying the cause of a new infectious outbreak to prevent its spread or personalizing a treatment based on a patient’s genetic variants to knock out a stubborn disease, modern Big Data analytics has a major role to play.
By leveraging cloud, Apache™ Hadoop®, next-generation sequencers, and other technologies, life scientists potentially have a new, very powerful way to conduct innovative global-scale collaborative genomic analysis research that has not been possible before. With the right approach, there are great benefits that can be realized.
To illustrate the possibilities and benefits of using coordinated worldwide genomic analysis, Dell partnered with researchers at Ben-Gurion University of the Negev (BGU) to develop a global data analytics environment that spans across multiple clouds. This environment lets life sciences organizations analyze data from multiple heterogeneous sources while preserving privacy and security. The work conducted by this collaboration simulated a scenario that might be used by researchers and public health organizations to identify the early onset of outbreaks of infectious diseases. The approach could also help uncover new combinations of virulence factors that may characterize new diseases. Additionally, the methods used have applicability to new drug discovery and translational and personalized medicine.
Expanding on past accomplishments
In 2003, SARS (severe acute respiratory syndrome) was the first infectious outbreak where fast global collaborative genomic analysis was used to identify the cause of a disease. The effort was carried out by researchers in the U.S. and Canada who decoded the genome of the coronavirus to prove it was the cause of SARS.
The Dell and BGU simulated disease detection and identification scenario makes use of technological developments (the much lower cost of sequencing, the availability of greater computing power, the use of cloud for data sharing, etc.) to address some of the shortcomings of past efforts and enhance the outcome.
Specifically, some diseases are caused by the combination of virulence factors. They may all be present in one pathogen or across several pathogens in the same biome. There can also be geographical variations. This makes it very hard to identified root causes of a disease when pathogens are analyzed in isolation as has been the case in the past.
Addressing these issues requires sequencing entire micro-biomes from many samples gathered worldwide. The computational requirements for such an approach are enormous. A single facility would need a compute and storage infrastructure on a par with major government research labs or national supercomputing centers.
Dell and BGU simulated a scenario of distributed sequencing centers scattered worldwide, where each center sequences entire micro-biome samples. Each center analyzes the sequence reads generated against a set of known virulence factors. This is done to detect the combination of these factors causing diseases, allowing for near-real time diagnostic analysis and targeted treatment.
To carry out these operations in the different centers, Dell extended the Hadoop framework to orchestrate distributed and parallel computation across clusters scattered worldwide. This pushed computation as close as possible to the source of data, leveraging the principle of data locality at world-wide scale, while preserving data privacy.
Since one Hadoop instance is represented by a single elephant, Dell concluded that a set of Hadoop instances, scattered across the world, but working in tandem formed a World Wide Herd or WWH. This is the name Dell has given to its Hadoop extensions.
Using WWH, Dell wrote a distributed application where each one of a set of collaborating sequence centers calculates a profile of the virulence factors present in each of the micro-biome it sequenced and sends just these profiles to a center selected to do the global computation.
That center would then use bi-clustering to uncover common patterns of virulence factors among subsets of micro-biomes that could have been originally sampled in any part of the world.
This approach could allow researchers and public health organizations to potentially identify the early onset of outbreaks and also uncover new combinations of virulence factors that may characterize new diseases.
There are several biological advantages to this approach. The approach eliminates the time required to isolate a specific pathogen for analysis and for re-assembling the genomes of the individual microorganisms. Sequencing the entire biome lets researchers identify known and unknown combinations of virulence factors. And collecting samples independently world-wide helps ensure the detection of variants.
On the compute side, the approach uses local processing power to perform the biome sequence analysis. This reduces the need for a large centralized HPC environment. Additionally, the method overcomes the matter of data diversity. It can support all data sources and any data formats.
This investigative approach could be used as a next-generation outbreak surveillance system. It allows collaboration where different geographically dispersed groups simultaneously investigate different variants of a new disease. In addition, the WWH architecture has great applicability to pharmaceutical industry R&D efforts, which increasingly relies on a multi-disciplinary approach where geographically dispersed groups investigate different aspects of a disease or drug target using a wide variety of analysis algorithms on share data.
Learn more about modern genomic Big Data analytics